Volume 8, Issue 2 (7-2020)
Jorjani Biomed J 2020, 8(2): 58-72 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Bagheri F, Tarokh M J, Ziaratban M. Two-stage Skin Lesion Segmentation from Dermoscopic Images by Using Deep Neural Networks. Jorjani Biomed J 2020; 8 (2) :58-72
URL: http://goums.ac.ir/jorjanijournal/article-1-737-en.html
URL: http://goums.ac.ir/jorjanijournal/article-1-737-en.html
1- Department of Industrial Engineering, K. N. Toosi University of Technology, Pardis Street, Molla Sadra Ave, Tehran, Iran
2- Department of Industrial Engineering, K. N. Toosi University of Technology, Pardis Street, Molla Sadra Ave, Tehran, Iran , f.bagheri@email.kntu.ac.ir
3- Department of Electrical Engineering, Faculty of Engineering, Golestan University, Gorgan, Iran
2- Department of Industrial Engineering, K. N. Toosi University of Technology, Pardis Street, Molla Sadra Ave, Tehran, Iran , f.bagheri@email.kntu.ac.ir
3- Department of Electrical Engineering, Faculty of Engineering, Golestan University, Gorgan, Iran
Full-Text [PDF 1223 kb]
(2298 Downloads)
| Abstract (HTML) (7678 Views)

Table1. Performance of different DeepLab structures with various pre-trained networks as a backbone on ISBI 2017 dataset
Table 2. Performance of different Yolo v2 structures with various pre-trained networks as a backbone on ISBI 2017 dataset
Table 3. Effect of using different image modes in normalization and segmentation stages on ISBI 2017 dataset
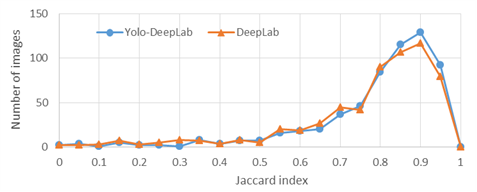
Figure 3. Distributions of the Jaccard index of test images on ISBI 2017 dataset
.png)
Figure 5. Normal images corresponding to the images in Figure 4. Green and red curves are boundaries of the correct lesions in the ground-truth and lesions segmented by the proposed method, respectively.
Conclusion
In this paper, a two-stage model for skin lesion segmentation is presented. First, the dermoscopic images were entered to the detection stage to estimate the rectangle area containing the skin lesion. The normal image were entered to another deep convolutional neural network with a DeepLab3+ architecture. The main motivation of using the detection stage before the segmentation stage was that the single-stage segmentation methods were generally sensitive to sizes and locations of lesions in dermoscopic images. Results in Figure 3 as well as the first and second row of Table 2 demonstrated the importance of existence of the detection stage before the segmentation. Experiments showed that the proposed skin lesion segmentation approach outperformed other methods on ISBI 2017 and DermQuest datasets.
Full-Text: (1617 Views)
Introduction
Cancer is one of the most common reasons of mortality in humans. One of the most prevalent cancers is melanoma cancer. This disease is started when melanocyte, a specific type of skin cell, starts to grow out of control. Based on the annual reports of American Cancer Society, melanoma causes the most rate of mortality in skin cancers. In 2020, about 100,350 new melanomas will be diagnosed (1). Therefore timely diagnosis of this disease is very important (2).
In recent decade, dermatologists have begun to use dermoscopy, one invasive imaging tool, to improve diagnosis of skin lesions of melanoma. This instrument provides an enlarged image of skin lesion through polarized light. It shows more details of skin structure, and improves the validity of the diagnosis (3). Diagnosis through large number of dermoscopy images by dermatologists is still difficult, time-consuming, and subjective. Thus, existence of automatic accurate skin lesion recognition systems is very helpful and even critical in timely diagnosis of skin cancers.
The first essential stage in any computer-based diagnostic systems is the object segmentation (4–6). The lesion segmentation is still as a challenge because of the large variety of skin lesions in shape, size, location, color, and texture. Existing additional factors such as hair, blood vessels, ruler sign, air bubbles, and also low contrast borders between lesions and surrounding tissues are amongst the obstacles of correct segmentation (7).
Generally, there are various methods for image segmentation, such as methods based on edge detection (8), thresholding (9), region detection (10), feature clustering (11), and also the methods based on deep neural networks (12). Recently segmentation of images by using deep learning and particularly, convolutional neural networks have achieved more accuracy in biomedical applications (13–18).
In 2017, Yu et al., developed an approach using deep residual networks (FCRN: fully convolutional residual network) for segmentation and classification of melanoma skin lesions. They tested their method on the ISBI 2016 dataset and achieved an accuracy of 94.9% (19). Lin et al., proposed two approaches for skin lesion segmentation, a method based on C-means clustering and a U-Net-based method. They evaluated their method on the ISBI 2017 dataset. The clustering-based technique and U-Net-based method achieved dice index of 61% and 77%, respectively (20). Yuan et al., offered a deep FCN-based skin lesion segmentation method. They used the Jaccard distance as a loss function and improved the basic FCN method. They examined their method on ISBI 2016 and PH2 datasets, and achieved accuracy of 95.5% and 93.8%, respectively (21). In 2017, Yuan et al., presented a skin lesion segmentation method using deep convolutional-deconvolutional neural networks (CDNN). They trained their model with different color spaces of dermoscopy images of the ISBI 2017 dataset. Their approach was ranked first in the ISBI 2017 Challenge achieving Jaccard Index of 76.5% (22).
Al-masni et al., at 2018 conducted a study on skin lesion segmentation and designed a full resolution convolutional network. They performed the examinations on ISBI 2017 and PH2 datasets and achieved the results of 77.11% and 84.79% by Jaccard criteria, respectively (7).
In our method, a two-stage CNN-based skin lesion segmentation method is proposed. Two different deep neural network structures is used in normalization and segmentation stages to improve the lesion segmentation performance.
Our contributions in this work are as follows:
- Proposing a two-stage segmentation method containing lesion detection stage before the segmentation stage.
- Employing robust deep neural architectures in both detection and segmentation stages.
- Use of 4 different modes of each input image in the detection stage and estimation of the bounding box based on the weighted averaging of the bounding boxes related to each mode.
- Use of 8 different modes of each normal image in the segmentation stage and constructing the final segmented image based on the segmentation results related to each mode.
Materials and Methods
Dataset
The proposed segmentation method will be evaluated on a well-known ISBI 2017 challenge dataset. This dataset was prepared by the International Skin Imaging Collaboration (ISIC) archive (23), and was presented online at (24). ISBI 2017 is the latest version of the datasets of dermoscopic images that contains segmentation ground-truth for all the training, test, and validation images. This dataset consists of 2750 8-bit RGB dermoscopy images of sizes from 540×722 to 4499×6748 pixels. A total of 2000, 150, and 600 images have been categorized for training, validation, and test, respectively.
To have a better evaluation, another dataset of non-dermoscopic images is used in our experiments. DermQuest consisted of 137 images (25).
Proposed method
In recent years, various single stage semantic segmentation methods such as U-net and FCN were used for medical image segmentation. The accuracy of these single-stage methods is sensitive to the size and location of the objects in images. Very large and very small lesions as well as various locations of lesions in images increase the complexity of trainable networks and reduce the performance. Therefore, it is better to perform a pre-segmentation step in order to normalize the size and location of lesions in images. It can reduce the complexity of the training procedure of the network in the segmentation stage and also increase the segmentation efficiency. In the proposed method, a detection stage is considered before the segmentation stage to normalize the size and location of a lesion in an input dermoscopy image. Figure 1 shows the framework of the proposed method.
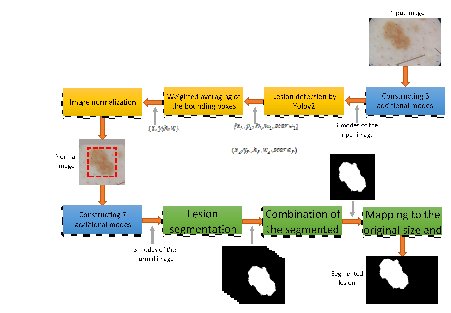
Figure 1. Framework of the proposed method
Cancer is one of the most common reasons of mortality in humans. One of the most prevalent cancers is melanoma cancer. This disease is started when melanocyte, a specific type of skin cell, starts to grow out of control. Based on the annual reports of American Cancer Society, melanoma causes the most rate of mortality in skin cancers. In 2020, about 100,350 new melanomas will be diagnosed (1). Therefore timely diagnosis of this disease is very important (2).
In recent decade, dermatologists have begun to use dermoscopy, one invasive imaging tool, to improve diagnosis of skin lesions of melanoma. This instrument provides an enlarged image of skin lesion through polarized light. It shows more details of skin structure, and improves the validity of the diagnosis (3). Diagnosis through large number of dermoscopy images by dermatologists is still difficult, time-consuming, and subjective. Thus, existence of automatic accurate skin lesion recognition systems is very helpful and even critical in timely diagnosis of skin cancers.
The first essential stage in any computer-based diagnostic systems is the object segmentation (4–6). The lesion segmentation is still as a challenge because of the large variety of skin lesions in shape, size, location, color, and texture. Existing additional factors such as hair, blood vessels, ruler sign, air bubbles, and also low contrast borders between lesions and surrounding tissues are amongst the obstacles of correct segmentation (7).
Generally, there are various methods for image segmentation, such as methods based on edge detection (8), thresholding (9), region detection (10), feature clustering (11), and also the methods based on deep neural networks (12). Recently segmentation of images by using deep learning and particularly, convolutional neural networks have achieved more accuracy in biomedical applications (13–18).
In 2017, Yu et al., developed an approach using deep residual networks (FCRN: fully convolutional residual network) for segmentation and classification of melanoma skin lesions. They tested their method on the ISBI 2016 dataset and achieved an accuracy of 94.9% (19). Lin et al., proposed two approaches for skin lesion segmentation, a method based on C-means clustering and a U-Net-based method. They evaluated their method on the ISBI 2017 dataset. The clustering-based technique and U-Net-based method achieved dice index of 61% and 77%, respectively (20). Yuan et al., offered a deep FCN-based skin lesion segmentation method. They used the Jaccard distance as a loss function and improved the basic FCN method. They examined their method on ISBI 2016 and PH2 datasets, and achieved accuracy of 95.5% and 93.8%, respectively (21). In 2017, Yuan et al., presented a skin lesion segmentation method using deep convolutional-deconvolutional neural networks (CDNN). They trained their model with different color spaces of dermoscopy images of the ISBI 2017 dataset. Their approach was ranked first in the ISBI 2017 Challenge achieving Jaccard Index of 76.5% (22).
Al-masni et al., at 2018 conducted a study on skin lesion segmentation and designed a full resolution convolutional network. They performed the examinations on ISBI 2017 and PH2 datasets and achieved the results of 77.11% and 84.79% by Jaccard criteria, respectively (7).
In our method, a two-stage CNN-based skin lesion segmentation method is proposed. Two different deep neural network structures is used in normalization and segmentation stages to improve the lesion segmentation performance.
Our contributions in this work are as follows:
- Proposing a two-stage segmentation method containing lesion detection stage before the segmentation stage.
- Employing robust deep neural architectures in both detection and segmentation stages.
- Use of 4 different modes of each input image in the detection stage and estimation of the bounding box based on the weighted averaging of the bounding boxes related to each mode.
- Use of 8 different modes of each normal image in the segmentation stage and constructing the final segmented image based on the segmentation results related to each mode.
Materials and Methods
Dataset
The proposed segmentation method will be evaluated on a well-known ISBI 2017 challenge dataset. This dataset was prepared by the International Skin Imaging Collaboration (ISIC) archive (23), and was presented online at (24). ISBI 2017 is the latest version of the datasets of dermoscopic images that contains segmentation ground-truth for all the training, test, and validation images. This dataset consists of 2750 8-bit RGB dermoscopy images of sizes from 540×722 to 4499×6748 pixels. A total of 2000, 150, and 600 images have been categorized for training, validation, and test, respectively.
To have a better evaluation, another dataset of non-dermoscopic images is used in our experiments. DermQuest consisted of 137 images (25).
Proposed method
In recent years, various single stage semantic segmentation methods such as U-net and FCN were used for medical image segmentation. The accuracy of these single-stage methods is sensitive to the size and location of the objects in images. Very large and very small lesions as well as various locations of lesions in images increase the complexity of trainable networks and reduce the performance. Therefore, it is better to perform a pre-segmentation step in order to normalize the size and location of lesions in images. It can reduce the complexity of the training procedure of the network in the segmentation stage and also increase the segmentation efficiency. In the proposed method, a detection stage is considered before the segmentation stage to normalize the size and location of a lesion in an input dermoscopy image. Figure 1 shows the framework of the proposed method.

Figure 1. Framework of the proposed method
Detection stage
The most important part of the proposed method is the estimation of the bounding box of the lesion in an image. Because the results of this stage substantially affect the segmentation performance. Any error in the detection stage results in high costs in the segmentation stage. Therefore, the accuracy of the detection stage is very important. We use object detection networks in our detection stage. Several methods based on deep convolutional neural networks have been proposed for object detection applications, such as R-CNN (11), Fast R-CNN (26), Faster R-CNN (27), Mask R-CNN (28), Single Shot multi-box detector (SSD) (29), You Only Look Once (Yolo) (30), and Retinanet (31).
Yolo is an object detection structure based on convolutional neural networks. This method divides the image into several sub-regions and for each region, predicts the boxes and their probabilities of belonging to the classes (30). Yolov2 structure is used in our detection stage to estimate the bounding box of the lesion in a dermoscopic image. The output of the Yolo is the coordinate and size of the bounding box of the detected lesion, as well as the detection score. In Figure. 1, x and y are coordinates of the left top corner of the box around the detected lesion. h and w are respectively the height and width of the bounding box, and score is the lesion detection score. The normalized image is constructed from the input image based on the location and size of the bounding box of the lesion detected by the Yolo. As shown in Figure. 1, the output of the detection stage is a normal image, in which the detected lesion is in the center. In this figure, red dashed rectangle is the bounding box of the lesion detected by Yolo. The Yolo v2 architecture requires a convolutional network as its backbone. We use some pre trained networks as its backbone and select the best one for our application.
To improve the accuracy of the detection stage, in addition to the original input image, three additional images are created by using rotation and flipping the input image. Each of these modes of the image is applied to the Yolo network and its corresponding bounding box of the lesion is estimated. Four modes of an input image are considered as follows: Input image, horizontal and vertical flips of input image, input image rotated by 180 degrees. The bounding box of each mode is flipped or rotated back to the original mode. The size and coordinate of the final box of the lesion is respectively calculated by applying weighted averaging on the sizes and coordinates of these four boxes estimated by Yolo. In the averaging operation, the weights are the scores of the detection results which are obtained at the output of the Yolo. Figure 2 shows the estimated bounding boxes related to four modes of an input image as well as the final bounding box calculated based on the weighted average of them.
The most important part of the proposed method is the estimation of the bounding box of the lesion in an image. Because the results of this stage substantially affect the segmentation performance. Any error in the detection stage results in high costs in the segmentation stage. Therefore, the accuracy of the detection stage is very important. We use object detection networks in our detection stage. Several methods based on deep convolutional neural networks have been proposed for object detection applications, such as R-CNN (11), Fast R-CNN (26), Faster R-CNN (27), Mask R-CNN (28), Single Shot multi-box detector (SSD) (29), You Only Look Once (Yolo) (30), and Retinanet (31).
Yolo is an object detection structure based on convolutional neural networks. This method divides the image into several sub-regions and for each region, predicts the boxes and their probabilities of belonging to the classes (30). Yolov2 structure is used in our detection stage to estimate the bounding box of the lesion in a dermoscopic image. The output of the Yolo is the coordinate and size of the bounding box of the detected lesion, as well as the detection score. In Figure. 1, x and y are coordinates of the left top corner of the box around the detected lesion. h and w are respectively the height and width of the bounding box, and score is the lesion detection score. The normalized image is constructed from the input image based on the location and size of the bounding box of the lesion detected by the Yolo. As shown in Figure. 1, the output of the detection stage is a normal image, in which the detected lesion is in the center. In this figure, red dashed rectangle is the bounding box of the lesion detected by Yolo. The Yolo v2 architecture requires a convolutional network as its backbone. We use some pre trained networks as its backbone and select the best one for our application.
To improve the accuracy of the detection stage, in addition to the original input image, three additional images are created by using rotation and flipping the input image. Each of these modes of the image is applied to the Yolo network and its corresponding bounding box of the lesion is estimated. Four modes of an input image are considered as follows: Input image, horizontal and vertical flips of input image, input image rotated by 180 degrees. The bounding box of each mode is flipped or rotated back to the original mode. The size and coordinate of the final box of the lesion is respectively calculated by applying weighted averaging on the sizes and coordinates of these four boxes estimated by Yolo. In the averaging operation, the weights are the scores of the detection results which are obtained at the output of the Yolo. Figure 2 shows the estimated bounding boxes related to four modes of an input image as well as the final bounding box calculated based on the weighted average of them.

Figure 2. Final bounding box estimation for an image of ISBI 2017 dataset. Red, green, blue, and yellow rectangles are the obtained bounding box related to 1st, 2nd, 3rd, and 4th modes of the input image, respectively. The black rectangle with dashed lines is the final bounding box calculated based on the weighted averaging of these 4 bounding boxes. The detection score is given with the corresponding color for each bounding box
Segmentation stage
The normalized image is delivered to the input of the segmentation stage. The output of this stage is a binary image in which the foreground pixels are the segmented lesion. Several methods and networks have been used for image segmentation in various applications. DeepLab architecture is one of the most recent structure which has performed well in many applications (32).
In general, the Deeplab architecture is based on a combination of two common architectures: Spatial Pyramid Pooling and Encoder-decoder networks (33). Different Deeplab structures have been improved over time. Deeplab V1 (32), Deeplab V2 (34), Deeplab V3 (35) and Deeplab V3+ (33), are the various structures of the Deeplab. In our segmentation stage, Deeplab V3+ is used to segment lesion from surrounding tissues. Several pre-trained network are considered as the backbone of the DeepLab and the best one is determined in our experiments.
In the segmentation stage, totally eight different modes of the normal image are considered to have more accurate segmentation result as follows: Input image, horizontal and vertical flips of the image, the image rotated by -45, 45, 90, 180, and 270 degrees. The output of each mode is flipped or rotated back to the original mode. The final binary result is obtained based on these eight binary images. A pixel in the final result is considered as foreground if it has nonzero value at least in 3 out of 8 result images.
Results
Evaluation metrics
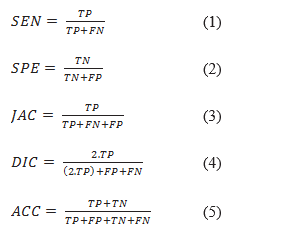
A commonly used metric to evaluate object detection methods is mean average precision (mAP). In addition, for evaluating segmentation methods, the following metrics have been used in the literature. Sensitivity (SEN) represents the rate of pixels of correctly detected skin lesion. Specificity (SPE) is the rate of pixels of non-skin lesions which are correctly classified (36). The Jaccard index (JAC) is an intersection over :union: (IOU) of the result of the segmented lesions with the ground truth masks (37). Index of Dice (DIC) measures the similarity of the segmented lesions through ground truth (16). Accuracy (ACC) shows the overall performance of the segmentation (37). All these criteria have been computed from four elements of the confusion matrix as follows:
In skin lesion segmentation applications, the main metric is the Jaccard index. The competitions on lesion segmentation, such as ISIC, have ranked the participants in terms of the Jaccard index.
Experimental setup
Deep neural networks require a lot of images in the training phase in order to adjust the training parameters. On the other hand, there are only 2000 dermoscopic images in the training set of the ISBI 2017 dataset. In order to increase the number of training images, data augmentation was performed by applying horizontal/vertical flipping, rotation, brightness changing, and resizing operations in a random manner on the images of the training set of the ISBI 2017 dataset. Finally a set of 14000 images were constructed and used in training of the Yolo network. These images are manually normalized and the normal images construct the training set for the DeepLab network.
In a similar way, the validation set of the ISBI 2017 dataset was augmented and a set of 600 images was constructed as our validation set. The augmented validation set were used to determine the best backbones for Yolo and DeepLab as well as preventing overfitting in the training phase of the DeepLab.
To perform experiments on the DermQuest dataset, images was randomly divided to two sets: training and test sets containing 103 and 34 images, respectively. Random division of images was done 4 times and the average results were reported in Table 5. The training images augmentation was done similar to the augmentation of the images in ISBI 2017 dataset. Finally, each augmented training set of the DermQuest consisted of 2163 images.
Experimental results
Several experiments were implemented to determine the proper backbone networks for Yolo v2 and DeepLab3+ architectures. A 6GB NVIDIA GeForce RTX2060 GPU were used in our experiments. The Initial learning rate and mini batch size were set to 0.001 and 8 samples, respectively. The learning rate was constant during Yolo training, and decreased by a factor of 0.3 every 10 epochs during training of the DeepLab. In Table 1, the Jaccard values obtained by the DeepLab with various backbones are reported. In this table, the detection stage was ignored and the input image was directly applied to the input of the DeepLab. Results on the augmented validation set showed that the best backbones for the Yolo and DeepLab architectures were vgg19 and Resnet101 networks, respectively. In subsequent experiments, the DeepLab based on Resnet101was used in the segmentation stage. Different pre-trained networks were used as a backbone of the Yolo v2 in the detection stage. The results are given in Table 2.
Effect of using more than one mode of images in normalization and segmentation stages is shown in Table 3. The proposed method is compared with various methods over the ISBI 2017 and DermQuest datasets based on the evaluation metrics in Table 4 and Table 5, respectively. It can be observed that the proposed lesion segmentation approach based on CNNs outperformed other methods.
The normalized image is delivered to the input of the segmentation stage. The output of this stage is a binary image in which the foreground pixels are the segmented lesion. Several methods and networks have been used for image segmentation in various applications. DeepLab architecture is one of the most recent structure which has performed well in many applications (32).
In general, the Deeplab architecture is based on a combination of two common architectures: Spatial Pyramid Pooling and Encoder-decoder networks (33). Different Deeplab structures have been improved over time. Deeplab V1 (32), Deeplab V2 (34), Deeplab V3 (35) and Deeplab V3+ (33), are the various structures of the Deeplab. In our segmentation stage, Deeplab V3+ is used to segment lesion from surrounding tissues. Several pre-trained network are considered as the backbone of the DeepLab and the best one is determined in our experiments.
In the segmentation stage, totally eight different modes of the normal image are considered to have more accurate segmentation result as follows: Input image, horizontal and vertical flips of the image, the image rotated by -45, 45, 90, 180, and 270 degrees. The output of each mode is flipped or rotated back to the original mode. The final binary result is obtained based on these eight binary images. A pixel in the final result is considered as foreground if it has nonzero value at least in 3 out of 8 result images.
Results
Evaluation metrics
A commonly used metric to evaluate object detection methods is mean average precision (mAP). In addition, for evaluating segmentation methods, the following metrics have been used in the literature. Sensitivity (SEN) represents the rate of pixels of correctly detected skin lesion. Specificity (SPE) is the rate of pixels of non-skin lesions which are correctly classified (36). The Jaccard index (JAC) is an intersection over :union: (IOU) of the result of the segmented lesions with the ground truth masks (37). Index of Dice (DIC) measures the similarity of the segmented lesions through ground truth (16). Accuracy (ACC) shows the overall performance of the segmentation (37). All these criteria have been computed from four elements of the confusion matrix as follows:

In skin lesion segmentation applications, the main metric is the Jaccard index. The competitions on lesion segmentation, such as ISIC, have ranked the participants in terms of the Jaccard index.
Experimental setup
Deep neural networks require a lot of images in the training phase in order to adjust the training parameters. On the other hand, there are only 2000 dermoscopic images in the training set of the ISBI 2017 dataset. In order to increase the number of training images, data augmentation was performed by applying horizontal/vertical flipping, rotation, brightness changing, and resizing operations in a random manner on the images of the training set of the ISBI 2017 dataset. Finally a set of 14000 images were constructed and used in training of the Yolo network. These images are manually normalized and the normal images construct the training set for the DeepLab network.
In a similar way, the validation set of the ISBI 2017 dataset was augmented and a set of 600 images was constructed as our validation set. The augmented validation set were used to determine the best backbones for Yolo and DeepLab as well as preventing overfitting in the training phase of the DeepLab.
To perform experiments on the DermQuest dataset, images was randomly divided to two sets: training and test sets containing 103 and 34 images, respectively. Random division of images was done 4 times and the average results were reported in Table 5. The training images augmentation was done similar to the augmentation of the images in ISBI 2017 dataset. Finally, each augmented training set of the DermQuest consisted of 2163 images.
Experimental results
Several experiments were implemented to determine the proper backbone networks for Yolo v2 and DeepLab3+ architectures. A 6GB NVIDIA GeForce RTX2060 GPU were used in our experiments. The Initial learning rate and mini batch size were set to 0.001 and 8 samples, respectively. The learning rate was constant during Yolo training, and decreased by a factor of 0.3 every 10 epochs during training of the DeepLab. In Table 1, the Jaccard values obtained by the DeepLab with various backbones are reported. In this table, the detection stage was ignored and the input image was directly applied to the input of the DeepLab. Results on the augmented validation set showed that the best backbones for the Yolo and DeepLab architectures were vgg19 and Resnet101 networks, respectively. In subsequent experiments, the DeepLab based on Resnet101was used in the segmentation stage. Different pre-trained networks were used as a backbone of the Yolo v2 in the detection stage. The results are given in Table 2.
Effect of using more than one mode of images in normalization and segmentation stages is shown in Table 3. The proposed method is compared with various methods over the ISBI 2017 and DermQuest datasets based on the evaluation metrics in Table 4 and Table 5, respectively. It can be observed that the proposed lesion segmentation approach based on CNNs outperformed other methods.
Table1. Performance of different DeepLab structures with various pre-trained networks as a backbone on ISBI 2017 dataset
| Backbone network of the DeepLab | Jaccard (%) | No. parameters (millions) |
| Resnet 101 | 77.02 | 44.6 |
| Resnet 50 | 76.84 | 25.6 |
| Vgg 19 | 76.72 | 144 |
| Vgg 16 | 75.56 | 138 |
| Resnet 18 | 74.97 | 11.7 |
| Densenet 201 | 74.85 | 20 |
| Alexnet | 74.82 | 61 |
| Mobilenet v2 | 74.78 | 3.5 |
| Googlenet | 74.18 | 7 |
| Squeezenet | 69.87 | 1.24 |
| Xception | 64.26 | 22.9 |
Table 2. Performance of different Yolo v2 structures with various pre-trained networks as a backbone on ISBI 2017 dataset
| Backbone network of the Yolo v2 | Jaccard (%) | |
| without detection stage | - | 77.02 |
| Vgg 19 | 91.23 | 79.05 |
| Resnet 101 | 90.51 | 78.86 |
| Vgg 16 | 91.47 | 78.73 |
| Resnet 50 | 90.54 | 78.72 |
| Resnet 18 | 90.66 | 78.36 |
| Densenet 201 | 90.49 | 78.34 |
| Mobilenet v2 | 88.84 | 78.01 |
| Alexnet | 88.81 | 77.69 |
| Googlenet | 84.53 | 76.24 |
| Squeezenet | 59.87 | 59.47 |
| Xception | 52.38 | 55.36 |
Table 3. Effect of using different image modes in normalization and segmentation stages on ISBI 2017 dataset
| Backbone of Yolo v2 | No. of image modes used in detection |
Backbone of DeepLab3+ | No. of image modes used in segmentation |
Jaccard (%) | |
| without detection stage | - | Resnet 101 | 1 | - | 76.24 |
| without detection stage | - | Resnet 101 | 8 | - | 77.02 |
| Vgg 19 | 1 | Resnet 101 | 1 | 90.07 | 77.69 |
| Vgg 19 | 1 | Resnet 101 | 8 | 90.07 | 78.43 |
| Vgg 19 | 4 | Resnet 101 | 1 | 91.23 | 78.36 |
| Vgg 19 | 4 | Resnet 101 | 8 | 91.23 | 79.05 |
Table 4. Quantitative comparison among various methods on ISBI 2017 dataset
| CDNN (22) | 82.50 | 97.50 | 93.40 | 84.90 | 76.50 |
| Li et al. (20) | 82.00 | 97.80 | 93.20 | 84.70 | 76.20 |
| ResNet (41) | 82.20 | 98.50 | 93.40 | 84.40 | 76.00 |
| U-Net (42) | - | - | - | 77.00 | 62.00 |
| FrCN (7) | 85.40 | 96.69 | 94.03 | 87.08 | 77.11 |
| DermoNet (43) | - | - | - | - | 78.30 |
| DSNet (44) | 87.5 | 95.5 | - | - | 77.5 |
| Proposed method | 88.90 | 95.47 | 94.11 | 87.13 | 79.05 |
Table 5. Quantitative comparison among various methods on the Dermquest dataset
| L-SRM (10) | 89.4 | 92.7 | 92.3 | 82.2 | 70.0 |
| Otsu-R (45) | 87.3 | 85.4 | 84.9 | 73.7 | 64.9 |
| Otsu-RGB (46) | 93.6 | 80.3 | 80.2 | 69.3 | 59.1 |
| Otsu-PCA (47) | 79.6 | 99.6 | 98.1 | 83.3 | 75.2 |
| TDLS (48) | 91.2 | 99.0 | 98.3 | 82.8 | 71.5 |
| Jafari (18) | 95.2 | 99.0 | 98.7 | 83.1 | 81.2 |
| FCN-8s (49) | 90.0 | 99.5 | 98.9 | 89.7 | 82.9 |
| U-net (50) | 91.5 | 99.5 | 98.7 | 88.7 | 81.4 |
| Yuan (21) | 91.6 | 99.6 | 98.7 | 89.3 | 82.0 |
| DFCN-No Interlace (51) | 78.0 | 99.8 | 98.0 | 81.62 | 73.0 |
| DFCN (51) | 92.4 | 99.6 | 98.9 | 91.6 | 85.2 |
| Proposed method | 95.14 | 99.50 | 99.20 | 92.30 | 85.92 |
Discussion
By comparing the first and second rows of Table 2, the impact of the detection stage before the segmentation is demonstrated. The main reason behind 2.03% improvement in Jaccard index is that the detection stage delivered normal images to the segmentation stage, in which the lesions had approximately similar sizes and located in the center. Therefore, the deep network in the segmentation stage faced the problem with lower complexity compared to the structure without the detection stage. In addition, results in Table 2 shows that the accuracy of the detection stage had a significant effect on the final Jaccard value. In other words, the Jaccard value was high when the mAP of the detection stage was high.
The distributions of Jaccard values of the test images are presented in Figure 3. The distribution has tended to the higher values of the Jaccard index by using the detection stage before the segmentation stage. Some segmentation results are shown in Figure 4. It can be observed that the results of the proposed two-stage method are more similar to the correct lesions compared to the single-stage method which did not contain the detection stage.
Results in Table 1 and Table 2 show that generally, Vgg and Resnet networks were more appropriate backbones compared to other pre-trained networks for Yolo and DeepLab architectures in the application of the skin lesion segmentation. It can be observed that the networks with very low number of parameters did not present high performance. Some pre-trained networks were not suitable as backbones of Yolo and DeepLab in this application because they were optimized for other specific applications. SqueezeNet was designed with very low number of parameters to make it possible to embed CNNs on Application-Specific Integrated Circuits (38) MobileNet was designed and optimized for mobile vision applications (39) . Xception contains depthwise separable convolutions to classify more complex classification problems. Xception presented small improvement in performance of classification of a 1000-classes problem on the ImageNet dataset. While it showed large gains on classifying 17000 different classes on the JFT dataset (40). But in the skin lesion application, there is only two classes. It may be a reason behind low performance of Xception in Table 1 and Table 2. We used additional image modes in normalization and segmentation stages. Table 3 shows that using additional image modes improved the performances of both detection and segmentation stages. The main reason was that due to the variety of sizes, locations, and shapes of lesions in dermoscopic images, for a test image, there may be more similar lesions in the training set to additional modes of the test image compared to its original mode. In other words, by using more modes of test image, more varieties of lesions were covered and caused the improvement of the performance.
By comparing the first and second rows of Table 2, the impact of the detection stage before the segmentation is demonstrated. The main reason behind 2.03% improvement in Jaccard index is that the detection stage delivered normal images to the segmentation stage, in which the lesions had approximately similar sizes and located in the center. Therefore, the deep network in the segmentation stage faced the problem with lower complexity compared to the structure without the detection stage. In addition, results in Table 2 shows that the accuracy of the detection stage had a significant effect on the final Jaccard value. In other words, the Jaccard value was high when the mAP of the detection stage was high.
The distributions of Jaccard values of the test images are presented in Figure 3. The distribution has tended to the higher values of the Jaccard index by using the detection stage before the segmentation stage. Some segmentation results are shown in Figure 4. It can be observed that the results of the proposed two-stage method are more similar to the correct lesions compared to the single-stage method which did not contain the detection stage.
Results in Table 1 and Table 2 show that generally, Vgg and Resnet networks were more appropriate backbones compared to other pre-trained networks for Yolo and DeepLab architectures in the application of the skin lesion segmentation. It can be observed that the networks with very low number of parameters did not present high performance. Some pre-trained networks were not suitable as backbones of Yolo and DeepLab in this application because they were optimized for other specific applications. SqueezeNet was designed with very low number of parameters to make it possible to embed CNNs on Application-Specific Integrated Circuits (38) MobileNet was designed and optimized for mobile vision applications (39) . Xception contains depthwise separable convolutions to classify more complex classification problems. Xception presented small improvement in performance of classification of a 1000-classes problem on the ImageNet dataset. While it showed large gains on classifying 17000 different classes on the JFT dataset (40). But in the skin lesion application, there is only two classes. It may be a reason behind low performance of Xception in Table 1 and Table 2. We used additional image modes in normalization and segmentation stages. Table 3 shows that using additional image modes improved the performances of both detection and segmentation stages. The main reason was that due to the variety of sizes, locations, and shapes of lesions in dermoscopic images, for a test image, there may be more similar lesions in the training set to additional modes of the test image compared to its original mode. In other words, by using more modes of test image, more varieties of lesions were covered and caused the improvement of the performance.

Figure 3. Distributions of the Jaccard index of test images on ISBI 2017 dataset
| (m) | (n) | (o) | (p) |
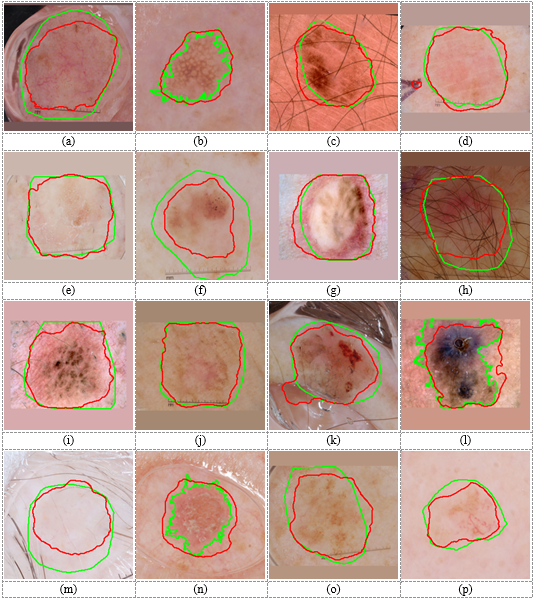
Figure 4. Lesion segmentation results on the ISBI 2017 dataset. Green curves are boundaries of correct lesions in the ground-truth made by experts. Blue and red curves are boundaries of lesions segmented by single-stage (DeepLab without normalization) and two-stage (Yolo-DeepLab) methods, respectively.
Figure 4 shows some dermoscopic images from ISBI 2017 dataset. It can be observed that by using the detection stage before the segmentation stage, the boundary of the segmented lesion will be more close to the correct boundary. In the detection-free approach, the DeepLab had to segment lesions with wide variety in location and size. Hence, DeepLab converged to the more frequent location and size of lesions in the training set. Therefore, the segmented result of the DeepLab in the detection-free approach for a very small lesion was bigger than the correct one (e.g., Figure 4 (b, k, n)). Also, it segmented smaller regions for very big lesions (e.g., Figure 4 (a, e, g)). In the two-stage approach, the DeepLab segmented lesions from normal images. Figure 5 depicts normal images corresponding to the images in Figure 4. As this figure shows, correct lesions in normal images are near the center and have similar sizes. Hence, the DeepLab segmented lesions with lower complexity and higher performance compared to the detection-free approach. Results in Table 4 and Table 5 demonstrate that the proposed is a dataset independent approach. Some segmentation results on images of the DermQuest dataset are shown in Figure 6.
.png)
Figure 5. Normal images corresponding to the images in Figure 4. Green and red curves are boundaries of the correct lesions in the ground-truth and lesions segmented by the proposed method, respectively.

Figure 6. Lesion segmentation results on the DermQuest dataset. Green and red curves are boundaries of correct lesions in the ground-truth and lesions segmented by the proposed method, respectively.
Conclusion
In this paper, a two-stage model for skin lesion segmentation is presented. First, the dermoscopic images were entered to the detection stage to estimate the rectangle area containing the skin lesion. The normal image were entered to another deep convolutional neural network with a DeepLab3+ architecture. The main motivation of using the detection stage before the segmentation stage was that the single-stage segmentation methods were generally sensitive to sizes and locations of lesions in dermoscopic images. Results in Figure 3 as well as the first and second row of Table 2 demonstrated the importance of existence of the detection stage before the segmentation. Experiments showed that the proposed skin lesion segmentation approach outperformed other methods on ISBI 2017 and DermQuest datasets.
|
Type of Article: Original article |
Subject:
Bio-statistics
Received: 2020/01/1 | Accepted: 2020/05/31 | Published: 2020/07/1
Received: 2020/01/1 | Accepted: 2020/05/31 | Published: 2020/07/1
References
1. National Cancer Institute. SEER Cancer Stat Facts: Melanoma of the Skin [Internet]. 2017 [cited 2020 Jul 15]. Available from: seer.cancer.gov › statfacts › html › melan [Google Scholar]
2. Balch CM, Gershenwald JE, Soong S-J, Thompson JF, Atkins MB, Byrd DR, et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol. 2009;27 (36):6199-6206. [view at publisher] [DOI] [Google Scholar]
3. Binder M, Schwarz M, Winkler A, Steiner A, Kaider A, Wolff KH. Pehamberger, Epiluminescence microscopy: a useful tool for the diagnosis of pigmented skin lesions for formally trained dermatologists. Arch Dermatol. 1995;131 (3):286-291.
https://doi.org/10.1001/archderm.131.3.286 [view at publisher] [DOI] [Google Scholar]
4. Celebi ME, Iyatomi H, Schaefer G, Stoecker WV. Lesion border detection in dermoscopy images. Comput Med Imaging Graph. 2009;33 (2):148-153. [view at publisher] [DOI] [Google Scholar]
5. Ganster H, Pinz P, Rohrer R, Wildling E, Binder M, Kittler H. Automated melanoma recognition. IEEE Trans Med Imaging. 2001;20 (3):233-239. [view at publisher] [DOI] [Google Scholar]
6. Celebi ME, Wen Q, Iyatomi H, Shimizu K, Zhou H, Schaefer G. A state-of-the-art survey on lesion border detection in dermoscopy images. Dermoscopy Image Anal. 2015;97-129. [view at publisher] [DOI] [Google Scholar]
7. Al-masni MA, Al-antari MA, Choi M, Han S, Kim T. Skin lesion segmentation in dermoscopy images via deep full resolution convolutional networks. Comput Methods Programs Biomed. 2018;162:221-31. [view at publisher] [DOI] [Google Scholar]
8. Kwak N-J, Kwon D-J, Kim Y-G, Ahn J-H. Color image segmentation using edge and adaptive threshold value based on the image characteristics. Proc 2004 Int Symp Intell Signal Process Commun Syst 2004 ISPACS 2004. 2004;555-558. [view at publisher] [Google Scholar]
9. Tan KS, Isa NAM. Color image segmentation using histogram thresholding fuzzy c-means hybrid approach. Pattern Recognit. 2011;44 (1):1-15. [view at publisher] [DOI] [Google Scholar]
10. Celebi ME, Kingravi HA, Iyatomi H, Aslandogan YA, Stoecker W V., Moss RH, et al. Border detection in dermoscopy images using statistical region merging. Ski Res Technol. 2008;14(3):347-53. [view at publisher] [DOI] [Google Scholar]
11. Girshick R, Donahue J, Darrell T, Malik J. Rich feature hierarchies for accurate object detection and semantic segmentation. Proc IEEE Conf Comput Vis Pattern Recognit. 2014;580-587. [view at publisher] [DOI] [Google Scholar]
12. Liu X, Deng Z, Yang Y. Recent progress in semantic image segmentation. Artif Intell Rev [Internet]. 2019;52(2):1089-106. Available from: [view at publisher] [DOI] [Google Scholar]
13. Yan Z, Zhan Y, Peng Z, Liao S, Shinagawa Y, Zhang S. Multi-instance deep learning: Discover discriminative local anatomies for bodypart recognition. IEEE Trans Med Imag. 2016;35 (5):1332-1343. [view at publisher] [DOI] [Google Scholar]
14. Dou Q, Chen H, Yu L, Zhao L, Qin J, Wang D. Automatic detection of cerebral microbleeds from MR images via 3D convolutional neural networks. IEEE Trans Med Imag. 2016;35 (5):1182-1195. [view at publisher] [DOI] [Google Scholar]
15. Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542 (7639):115-118. [view at publisher] [DOI] [Google Scholar]
16. Pereira S, Pinto A, Alves V, Silva CA. Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans Med Imaging. 2016;35 (5):1240-1251. [view at publisher] [DOI] [Google Scholar]
17. Avendi M, Kheradvar A, Jafarkhani H. A combined deep-learning and deformable-model approach to fully automatic segmentation of the left ventricle in cardiac MRI. Med Image Anal. 2016;30:108-119. [view at publisher] [DOI] [Google Scholar]
18. Jafari MH, Nasr-Esfahani E, Karimi N, Soroushmehr S, Samavi S, Najarian K. Extraction of skin lesions from non-dermoscopic images using deep learning. Int J Comput Assist Radiol Surg. 2017;12(6):1021-30. [view at publisher] [DOI] [Google Scholar]
19. Yu L, Chen H, Dou Q, Qin J, Heng P-A. Automated melanoma recognition in dermoscopy images via very deep residual networks. IEEE Trans Med Imaging. 2017;36 (4):994-1004. [view at publisher] [DOI] [Google Scholar]
20. Li Y, Shen L. Skin lesion analysis towards melanoma detection using deep learning network. Sensors. 2018;18(2 (556)):1-16. [view at publisher] [DOI] [Google Scholar]
21. Yuan Y, Chao M, Lo Y-C. Automatic skin lesion segmentation using deep fully convolutional networks with Jaccard distance. IEEE Trans Med Imaging. 2017;36(9):1876-1886. [view at publisher] [DOI] [Google Scholar]
22. Yuan Y, Chao M, Lo Y-C. Automatic skin lesion segmentation with fully convolutional-deconvolutional networks. IEEE Trans Med Imaging. 2017;36 (9):1876-1886. [DOI] [Google Scholar]
23. Codella NCF, Gutman D, Celebi ME, Helba B, Marchetti MA, Dusza SW, et al. Skin Lesion Analysis Toward Melanoma Detection: a Challenge at The 2017 International Symposium on Biomedical Imaging (ISBI), Hosted By The International Skin Imaging Collaboration (ISIC). arXiv:171005006v3. 2017; [view at publisher] [DOI] [Google Scholar]
24. ISIC, 2017. Skin lesion analysis towards melanoma detection. available [Accessed 08 02 2018] https//challenge.kitware.com/#challenge/n/%0AISIC_2017%3A_Skin_Lesion_Analysis_Towards_Melanoma_Detection [Internet]. Available from: available: [Accessed 08 02 2018] https://challenge.kitware.com/#challenge/n/%0AISIC_2017%3A_Skin_Lesion_Analysis_Towards_Melanoma_Detection [Google Scholar]
25. DermQuest. The art, science and practice of dermatology. http://www.dermquest.com. 2010; [Google Scholar]
26. Girshick R. Fast R-CNN. Proc IEEE Int Conf Comput Vis. 2015;1440-1448. [view at publisher] [DOI] [Google Scholar]
27. Ren S, He K, Girshick R, Sun J. Faster R-CNN : Towards Real-Time Object Detection with Region Proposal Networks. Adv Neural Inf Process Syst. 2015;91-99. [view at publisher] [Google Scholar]
28. He K, Gkioxari G, Dollár P, Girshick R. Mask R-CNN. IEEE Trans Pattern Anal Mach Intell. 2020;42(2):386-97. [view at publisher] [DOI] [Google Scholar]
29. Liu W, Anguelov D, Erhan D, Szegedy C, Reed S, Fu C, et al. SSD : Single Shot MultiBox Detector. Comput Vis - ECCV 2016 Lect Notes Comput Sci vol 9905 Springer, Cham. 2016; [view at publisher] [DOI] [Google Scholar]
30. Redmon J, Divvala S, Girshick R, Farhadi A. You Only Look Once: Unified, Real-Time Object Detection. Proc IEEE Conf Comput Vis Pattern Recognit. 2016;779-788. [view at publisher] [DOI] [Google Scholar]
31. Lin TY, Goyal P, Girshick R, He K, Dollar P. Focal Loss for Dense Object Detection. arXiv:170802002v2. 2018; [view at publisher] [DOI] [Google Scholar]
32. Chen L-C, Papandreou G, Kokkinos I, Murphy K, Yuille AL. Semantic Image Segmentation with Deep Convolutional Nets and Fully Connected CRFs. ICLR, 2015. 2015; [view at publisher] [Google Scholar]
33. Chen L, Zhu Y, Papandreou G, Schroff F, Adam H. Encoder-Decoder with Atrous Separable Convolution for Semantic Image Segmentation. arXiv:180202611v3. 2018; [view at publisher] [DOI] [Google Scholar]
34. Chen LC, Papandreou G, Kokkinos I, Murphy K, Yuille AL. DeepLab: Semantic Image Segmentation with Deep Convolutional Nets, Atrous Convolution, and Fully Connected CRFs. IEEE Trans Pattern Anal Mach Intell. 2017;40(4):834-48. [view at publisher] [DOI] [Google Scholar]
35. Chen L-C, Papandreou G, Schroff F, Adam H. Rethinking Atrous Convolution for Semantic Image Segmentation. http://arxiv.org/abs/170605587 [Internet]. 2017; Available from: http://arxiv.org/abs/1706.05587 [view at publisher] [Google Scholar]
36. Al-antari MA, Al-masni MA, Park SU, Park JH, Metwally MK, Kadah YM, et al. An automatic computer-aided diagnosis system for breast cancer in digital mammograms via deep belief network. J Med Biol Eng. 2017;1-14. [view at publisher] [DOI] [Google Scholar]
37. Powers D. Evaluation: from precision, recall and F-measure to ROC, informedness, markedness and correlation. J Mach Learn Technol. 2011;2 (1):37-63. [view at publisher] [Google Scholar]
38. Iandola FN, Han S, Moskewicz MW, Ashraf K, Dally WJ, Keutzer K. SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and <0.5MB model size. arXiv:160207360v4 [Internet]. 2016;1-13. Available from: http://arxiv.org/abs/1602.07360 [view at publisher] [Google Scholar]
39. Howard AG, Zhu M, Chen B, Kalenichenko D, Wang W, Weyand T, et al. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. 2017; Available from: http://arxiv.org/abs/1704.04861 [view at publisher] [Google Scholar]
40. Chollet F. Xception: Deep learning with depthwise separable convolutions. Proc - 30th IEEE Conf Comput Vis Pattern Recognition, CVPR 2017. 2017;2017-Janua:1800-7. [view at publisher] [DOI] [Google Scholar]
41. Bi L, Kim J, Ahn E, Feng D. Automatic Skin Lesion Analysis using Large-scale Dermoscopy Images and Deep Residual Networks. http://arxiv.org/abs/170304197 [Internet]. 2017;6-9. Available from: http://arxiv.org/abs/1703.04197 [view at publisher] [Google Scholar]
42. Lin BS, Michael K, Kalra S, Tizhoosh HR. Skin lesion segmentation: UNets versus clustering. arXiv: 171001248. 2017; [view at publisher] [DOI] [Google Scholar]
43. Baghersalimi S, Bozorgtabar B, Schmid-saugeon P, Ekenel HK, Thiran J. DermoNet : densely linked convolutional neural network for efficient skin lesion segmentation. EURASIP J Image Video Process. 2019;71:1-10. [view at publisher] [DOI] [Google Scholar]
44. Hasan MK, Dahal L, Samarakoon PN, Tushar FI, Martí R. DSNet: Automatic dermoscopic skin lesion segmentation. Comput Biol Med. 2020;120(April):103738. [view at publisher] [DOI] [Google Scholar]
45. Cavalcanti PG, Yari Y, Scharcanski J. Pigmented Skin Lesion Segmentation on Macroscopic Images Review of Recent Pigmented Skin Lesion Segmentation Methods. Proceeding ICNZ. 2010; [view at publisher] [DOI] [Google Scholar]
46. Cavalcanti PG, Scharcanski J, Lopes CBO. Shading Attenuation in Human Skin Color Images. Adv Vis Comput Lect Notes Comput Sci. 2010;6453:190-8. [view at publisher] [DOI] [Google Scholar]
47. Cavalcanti PG, Scharcanski J. Automated prescreening of pigmented skin lesions using standard cameras. Comput Med Imaging Graph. 2011;35(6):481-91. [view at publisher] [DOI] [Google Scholar]
48. Glaister J, Member S, Wong A, Clausi D a, Member S. Segmentation of Skin Lesions From Digital Images Using Joint Statistical Texture Distinctiveness. IEEE Trans Biomed Eng. 2014;61(4):1220-30. [view at publisher] [DOI] [Google Scholar]
49. Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. Proc CVPR. 2015;3431-40. [view at publisher] [DOI] [Google Scholar]
50. Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. Int Conf Med Image Comput Comput Interv. 2015;234-241. [view at publisher] [DOI] [Google Scholar]
51. Nasr-Esfahani E, Rafiei S, Jafari MH, Karimi N, Wrobel JS, Soroushmehr SMR, et al. Dense pooling layers in fully convolutional network for skin lesion segmentation. Comput Med Imaging Graph [Internet]. 2019;78:101658. Available from: http://arxiv.org/abs/1712.10207. [view at publisher] [DOI] [Google Scholar]
Send email to the article author
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |






